What is this fundamental unit of microbial diversity, and why is it crucial for understanding the microbial world?
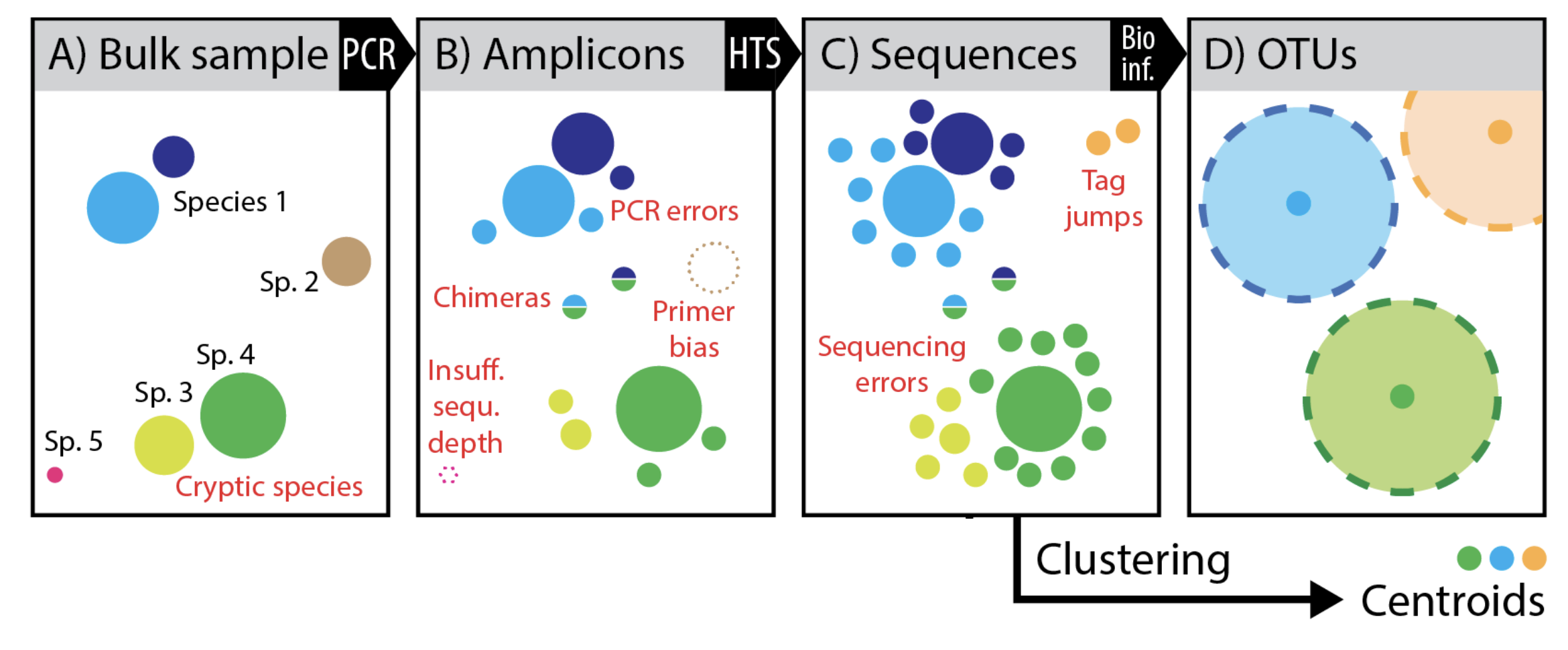
This taxonomic unit represents a cluster of closely related microorganisms. It is typically defined based on a comparison of their genetic sequences, such as ribosomal RNA genes. Consequently, sequences sharing a high degree of similarity are grouped into the same operational taxonomic unit. For example, different bacterial strains isolated from the same environmental sample may belong to the same OTU if their genetic sequences are very similar.
Operational taxonomic units (OTUs) are fundamental to ecological studies of microbes because they facilitate classifying and quantifying microbial diversity in complex environments like soil, water, or the human gut. By grouping similar microbial sequences, researchers can determine the composition of microbial communities, track changes in community structure over time, and potentially link this structure to environmental factors. This allows researchers to address crucial ecological and evolutionary questions about microbial communities and their interactions.
Moving forward, this understanding of microbial communities, as represented by OTUs, is pivotal to fields ranging from medicine to agriculture to environmental science. Understanding these microbial communities is becoming increasingly important for advancing various fields.
OTU
Operational Taxonomic Units (OTUs) are fundamental to microbial ecology, serving as a cornerstone for understanding microbial diversity and community composition. Their significance lies in simplifying complex microbial communities into manageable units.
- Microbial diversity
- Community analysis
- Genetic similarity
- Taxonomic classification
- Environmental factors
- Sequence-based identification
- Comparative genomics
- Ecological relationships
OTUs facilitate the study of microbial communities by classifying microorganisms based on shared genetic sequences (e.g., 16S rRNA). Analyzing OTU composition in different environments (soil, water, human gut) reveals how environmental factors influence microbial communities. Comparative genomics within OTUs informs evolutionary relationships. Understanding these ecological relationships is essential for predicting community responses to changing environments. For instance, variations in OTU composition across soil samples can correlate with nutrient levels, demonstrating the link between environmental conditions and microbial communities.
1. Microbial Diversity
Microbial diversity encompasses the vast array of microorganisms inhabiting diverse environments. Quantifying and understanding this diversity are crucial for comprehending ecosystem function and dynamics. Operational Taxonomic Units (OTUs) play a pivotal role in this process by providing a framework for classifying and analyzing microbial communities. OTUs, defined based on genetic similarities, allow researchers to characterize and compare microbial diversity across various samples.
- Classification and Quantification
OTUs enable the classification of microorganisms into groups based on shared genetic sequences. This process facilitates the quantification of microbial diversity within a sample by identifying the number and abundance of different OTUs. For instance, comparing OTU profiles from a pristine forest soil versus a contaminated one reveals the impact of pollution on the microbial community composition.
- Community Structure and Function
Analyzing OTU composition reveals insights into microbial community structure and function. Variations in OTU abundance between different environments highlight the adaptation of microorganisms to specific conditions. Moreover, the presence or absence of particular OTUs can indicate the activity or influence of specific metabolic pathways, crucial for comprehending ecosystem services like nutrient cycling.
- Environmental Impacts
Changes in microbial diversity, as reflected by OTU profiles, are sensitive indicators of environmental alterations. Comparing OTU composition in a polluted stream to a reference stream allows for assessing the impact of pollution on aquatic microbial communities. This method is essential in environmental monitoring and management.
- Evolutionary Relationships
Analyzing OTU composition across different environments helps track evolutionary relationships among microbial species. By comparing the evolutionary history of microorganisms across space and time, researchers develop a better understanding of their adaptation to varying environmental conditions. Consequently, this knowledge is useful for predicting responses to future environmental changes.
In conclusion, microbial diversity, assessed through OTUs, provides valuable information regarding community structure and function, environmental impacts, and evolutionary relationships. This information is vital for ecological studies, environmental monitoring, and predicting responses to external stimuli like pollution or climate change. Furthermore, it serves as a vital tool in understanding the intricate roles of microorganisms in complex ecosystems.
2. Community analysis
Community analysis, a crucial aspect of ecological research, hinges on the identification and quantification of diverse species within a particular area. Operational Taxonomic Units (OTUs) play a fundamental role in this process. OTUs, defined by shared genetic characteristics, serve as proxies for species or groups of closely related microorganisms. Analysis of OTU composition within a community provides a means to understand the structure, function, and dynamics of that community. For example, comparing OTU profiles from different soil samples allows for the identification of distinct microbial communities responding to variations in environmental conditions, such as nutrient availability or pollution levels. Recognizing these patterns is vital for predicting the consequences of environmental perturbations.
The importance of community analysis in the context of OTUs extends to various applications. In environmental monitoring, for instance, tracking changes in OTU composition over time can reveal the impact of pollution or other environmental stressors on microbial ecosystems. This information is crucial for assessing ecosystem health and informing remediation strategies. In medical research, community analysis using OTUs can identify microbial communities associated with specific diseases. Understanding the microbial composition in a patient's gut, for example, may shed light on the causes and potential treatments of certain conditions. Further, in agricultural settings, community analysis employing OTUs allows for the identification of beneficial microbes that contribute to soil fertility, paving the way for sustainable agricultural practices. In all these cases, the ability to analyze communities through the lens of OTUs is pivotal for understanding ecological processes and improving human-environmental interactions.
In summary, community analysis, facilitated by OTUs, allows for the comprehensive understanding of microbial communities. This includes identifying and quantifying different species, analyzing their function, and tracking the effects of environmental changes. The practical applications of this understanding are extensive, reaching from environmental monitoring to medical research and agriculture. While challenges in determining precise species boundaries within OTUs remain, the value of community analysis employing OTUs as a tool for ecological research is undeniable.
3. Genetic Similarity
Genetic similarity forms the bedrock of operational taxonomic units (OTUs). OTUs group organisms based on shared genetic sequences, particularly in the case of microorganisms where traditional taxonomic methods may be insufficient. The degree of genetic similarity is directly correlated with the taxonomic classification. High genetic similarity indicates a closer evolutionary relationship between organisms, suggesting they belong to the same OTU. Conversely, low genetic similarity implies a distant evolutionary connection and likely membership in different OTUs. This relationship is fundamental to understanding the diversity and structure of microbial communities.
The concept of genetic similarity is crucial for characterizing microbial communities in various environments. Comparing DNA sequences, such as those of the 16S rRNA gene in bacteria, allows researchers to identify OTUs. For instance, in analyzing soil samples, comparing the genetic similarity of microbial communities exposed to different types of pollution reveals how pollution impacts community structure. High similarity within an OTU suggests that organisms share a common ancestor and likely possess similar metabolic capabilities. This can be critical in assessing ecosystem functionality, as organisms within the same OTU may contribute similarly to nutrient cycles or other vital processes. Similarly, in medical contexts, identifying microbial OTUs based on genetic similarity in a patient's gut can help determine if specific pathogens or beneficial bacteria are present, aiding in diagnosis and treatment planning.
In essence, genetic similarity serves as a defining characteristic for OTUs, enabling the classification and analysis of diverse microbial communities across different environments and contexts. While challenges in interpreting the precise boundaries of species based solely on genetic similarity exist, understanding the relationship between genetic similarity and OTUs is vital for comprehensive ecological studies, environmental monitoring, and medical research. This understanding facilitates the identification of microbial communities, their functions, and their responses to environmental factors, offering valuable insights into the complexity and interconnectedness of life.
4. Taxonomic Classification
Taxonomic classification provides a hierarchical system for organizing and categorizing living organisms, including microorganisms. This system is essential for understanding the relationships between organisms and their diversity, and it directly impacts the concept of operational taxonomic units (OTUs). The hierarchical structure of classification, ranging from broad domains to specific species, informs the grouping and characterization of organisms within OTUs.
- Hierarchical Structure and OTUs
Taxonomic classification organizes life into a hierarchy of nested groups, from domains (e.g., Bacteria, Archaea, Eukarya) down to species. OTUs, in contrast, are operational groupings based on shared characteristics, often genetic similarity. The hierarchical structure of classification informs the interpretation of OTU groupings. For instance, if multiple OTUs share a common genus, this suggests a closer evolutionary relationship than OTUs belonging to different phyla. This understanding is vital in ecological analyses to evaluate the evolutionary and functional relationships between microorganisms within a given environment.
- Genetic Similarity and Classification
In the context of microorganisms, taxonomic classification increasingly relies on genetic similarity, particularly 16S rRNA gene sequencing. Similar genetic sequences indicate a closer evolutionary relationship, leading to placement within the same taxonomic groups, and consequently to the same OTUs. OTUs based on genetic data provide an essential tool for characterizing microbial communities, enabling comparisons and interpretations that further microbial ecology and medical research.
- Species Definition and OTU Boundaries
The concept of species can be complex and challenging to define precisely, especially for microorganisms. OTUs, however, provide operational definitions for grouping organisms based on shared genetic characteristics. These groups may or may not align perfectly with established species definitions, and the boundaries between OTUs can be refined and adjusted as new data emerge. This implies that a thorough understanding of evolutionary relationships and phylogenetic analyses is essential in the interpretation of OTU composition and diversity.
- Applications in Microbial Ecology
Taxonomic classification is fundamental to microbial ecology. Researchers use the established taxonomic framework to interpret OTU data. For instance, understanding the taxonomic affiliation of OTUs in a particular environment (soil, water) allows researchers to connect the observed microbial community composition with environmental variables, ecological functions, and potentially ecosystem services.
In conclusion, taxonomic classification provides a crucial framework for understanding the relationships between organisms and is directly relevant to the characterization of OTUs. The combination of taxonomic and genetic information provides a more complete picture of microbial diversity and community structure, facilitating crucial ecological interpretations. Further research in this area is essential to address ongoing challenges in microbial classification and refine our understanding of microbial biodiversity and its roles in different ecosystems.
5. Environmental Factors
Environmental factors exert a profound influence on microbial communities, significantly shaping their composition and diversity, as reflected in operational taxonomic units (OTUs). Changes in environmental conditions can lead to shifts in the relative abundance of different OTUs. For instance, variations in nutrient levels, temperature, pH, and salinity can drive community restructuring, altering the dominance of particular OTU groups. The impact of these factors is often observed in natural ecosystems. Polluted water bodies, for example, frequently exhibit distinct OTU profiles compared to unpolluted ones. This difference arises from the selective pressures exerted by pollutants on microbial populations, leading to the proliferation of specific OTUs adapted to the changed conditions.
Understanding the interplay between environmental factors and OTU composition is crucial for diverse applications. In environmental monitoring, tracking changes in OTU profiles provides early warning signals of environmental degradation. By identifying specific OTUs sensitive to particular stressors, scientists can assess the impact of pollution, climate change, or other environmental alterations on ecosystems. These insights can guide remediation efforts and conservation strategies. In agricultural contexts, manipulating environmental factors can be used to promote the growth of beneficial OTUs crucial for soil health and fertility. Optimized nutrient availability, for instance, can encourage specific OTUs with key roles in nutrient cycling, leading to more sustainable and productive agricultural systems. In medical contexts, identifying specific microbial OTUs linked to specific environmental exposures (e.g., those associated with water contamination) can inform public health initiatives and strategies for mitigating health risks.
In conclusion, environmental factors play a pivotal role in determining the structure and composition of microbial communities, as represented by OTUs. The influence of environmental conditions on OTU profiles is multifaceted and deeply interconnected. This understanding provides a powerful framework for environmental monitoring, ecological assessment, and management strategies across diverse fields. While the complexity of microbial interactions remains a challenge, recognizing the profound influence of environmental parameters on OTU composition offers significant potential for predicting and mitigating the effects of environmental change on biological systems.
6. Sequence-based identification
Sequence-based identification is a cornerstone method for characterizing and classifying microorganisms, particularly crucial for understanding operational taxonomic units (OTUs). This approach leverages the unique genetic code embedded within the DNA or RNA sequences of organisms to determine their taxonomic affiliations and relationships. This process is critical in microbial ecology for analyzing microbial community compositions, evaluating the impacts of environmental changes, and identifying novel or rare organisms.
- 16S rRNA gene sequencing
The 16S rRNA gene, highly conserved yet variable across different microbial species, serves as a primary target for sequence-based identification. This gene's sequence variation reflects phylogenetic relationships, allowing researchers to classify microorganisms and group them into OTUs based on shared ancestry. Using this method, the composition of microbial communities in various environments, from soil to water to the human gut, can be effectively analyzed. Furthermore, new species and strains can be discovered by identifying unique 16S rRNA sequences.
- DNA barcoding
Extending beyond the 16S rRNA gene, DNA barcoding utilizes specific marker genes from different parts of the genome. This approach provides a broader view of microbial diversity and can potentially discern relationships among OTUs not fully characterized by 16S rRNA sequences. For example, different genes might offer insights into the metabolic capabilities of microorganisms within an OTU, aiding ecological interpretations.
- Phylogenetic analysis
Sequence-based identification enables phylogenetic analyses to construct evolutionary trees representing the relationships among different OTUs. These analyses provide insights into the evolutionary history of microorganisms and their adaptation to various environmental conditions. Comparing the phylogenetic trees of OTUs from diverse environments can highlight adaptive strategies and evolutionary trends, thus contributing significantly to our understanding of microbial community dynamics.
- Computational tools
Advancements in computational biology have led to robust algorithms and databases for sequence analysis and comparison. These tools streamline the process of aligning and comparing sequences from different organisms. They also permit the identification of OTUs by comparing sequences against existing databases. This rapid and comprehensive analysis of data is essential for large-scale studies of microbial diversity and community structure.
In conclusion, sequence-based identification provides the foundational data for characterizing and classifying OTUs. The use of 16S rRNA gene sequencing, DNA barcoding, phylogenetic analysis, and powerful computational tools not only reveal the evolutionary relationships within microbial communities but also provide insights into their responses to environmental factors, contributing significantly to the understanding of microbial ecology and evolution. This robust approach underpins various applications, from environmental monitoring to biomedical research, emphasizing the critical role sequence-based methods play in uncovering the intricate nature of microbial ecosystems.
7. Comparative Genomics
Comparative genomics plays a crucial role in the characterization and understanding of operational taxonomic units (OTUs). By comparing the genomes of different organisms within an OTU, researchers gain insights into evolutionary relationships, functional capabilities, and adaptations to environmental conditions. This analysis identifies conserved genes and specific genetic variations, offering crucial data for the delineation and classification of OTUs. For instance, comparing the genomes of bacteria within a particular OTU associated with a specific soil type might reveal genes related to nutrient acquisition or stress tolerance, providing mechanistic details about their ecological role. The comparison of these genome sequences can also reveal gene losses or gains, which indicate evolutionary adaptations to specific environments.
The practical significance of this approach extends to various fields. In medicine, comparative genomics can identify pathogenic bacteria within a specific OTU, revealing specific virulence factors or antibiotic resistance mechanisms. This understanding can inform the development of targeted therapies. In environmental science, comparative genomics of microbial OTUs can elucidate how organisms adapt to pollutants or extreme conditions, offering valuable insights into the resilience of ecosystems. This information can aid in the prediction of community responses to environmental changes. Additionally, comparative genomics provides a deeper understanding of the evolutionary history of OTUs, illuminating the diversification and adaptation of these microbial lineages. By studying the genomic differences, researchers can refine the classification of OTUs, potentially resolving previously ambiguous taxonomic assignments, which ultimately enhances the precision of microbial community analyses.
In summary, comparative genomics provides a powerful tool for understanding OTUs. By analyzing genomic similarities and differences, researchers can gain valuable insights into the evolution, function, and adaptation of microbial communities. This detailed understanding has implications for diverse fields, including medicine, environmental science, and evolutionary biology. However, the computational demands of comparing large numbers of genomes can pose a challenge, and the precise interpretation of genomic data requires careful consideration of evolutionary context. Nevertheless, the use of comparative genomics strengthens our comprehension of OTUs, leading to more accurate and detailed analyses of microbial communities.
8. Ecological Relationships
Ecological relationships are integral to understanding operational taxonomic units (OTUs). OTUs represent groups of microorganisms with shared genetic characteristics. These shared characteristics often reflect similar ecological roles and interactions within a community. Strong ecological relationships between OTUs influence the abundance and distribution of those units. For instance, in a nutrient-poor soil environment, OTUs specialized in nutrient acquisition might be more prevalent and have stronger interactions compared to those with different metabolic needs. Similarly, in a symbiotic relationship, the presence of specific OTUs might be dependent on the presence of others, influencing the community's overall structure and function. A direct cause-and-effect relationship exists: specific environmental conditions select for certain OTUs with adaptations conducive to those conditions, thus shaping their interactions and their prominence within the community. This is exemplified in studies on the human gut microbiome, where certain OTUs are associated with specific dietary habits and can influence nutrient absorption.
Understanding the ecological relationships between OTUs is crucial for several applications. In agricultural settings, recognizing the beneficial interactions between specific OTUs and plant roots can lead to improved crop yields and reduced reliance on fertilizers. In environmental monitoring, observing changes in the interactions between OTUs can signal ecosystem stress and potential shifts in functionality. In medical contexts, investigating the ecological relationships of OTUs within the human gut can aid in understanding the complexities of conditions like inflammatory bowel disease and other microbiome-related disorders. Furthermore, studying the ecological dynamics among OTUs can contribute to predicting the responses of microbial communities to various perturbations and provide critical knowledge for conservation efforts. The ability to understand how OTUs interact with each other within an ecosystem is foundational to forecasting and managing the ecosystem's overall health.
In conclusion, ecological relationships are not separate from OTUs; they are fundamentally linked. Specific environmental pressures drive selection for certain OTUs and influence their interactions within a community. Analyzing these relationships reveals insights into the functional roles of microorganisms and their contributions to ecosystem processes. Further research into the intricate web of ecological relationships between OTUs is vital for managing and predicting the outcomes of environmental changes. This understanding has far-reaching implications for fields ranging from agriculture to medicine and environmental science, enabling effective interventions and strategies to support the health and resilience of diverse ecosystems.
Frequently Asked Questions about OTUs
This section addresses common queries regarding operational taxonomic units (OTUs). A clear understanding of OTUs is essential for interpreting microbial community data.
Question 1: What is an OTU, exactly?
An operational taxonomic unit (OTU) is a cluster of microorganisms sharing a high degree of similarity in a specific gene sequence, often 16S rRNA. This similarity indicates a close evolutionary relationship, but does not necessarily equate to a formally recognized taxonomic grouping like a species. OTUs provide a practical way to characterize complex microbial communities by grouping organisms with similar traits.
Question 2: How are OTUs identified?
OTUs are typically identified by comparing the sequences of specific genes, such as the 16S rRNA gene. Sequence similarity is used to determine which organisms are grouped together into the same OTU. Bioinformatic tools and statistical methods facilitate this process.
Question 3: What is the significance of OTUs in ecological studies?
OTUs are invaluable in ecological studies. By grouping microorganisms with similar characteristics, researchers can analyze community structure and diversity, assess the impact of environmental factors, and potentially understand the functional roles of different microbial groups.
Question 4: How are OTUs related to traditional taxonomic classifications?
OTUs are distinct from traditional taxonomic classifications. While OTU groupings often overlap with existing taxonomic categories, OTUs are operational groupings based on shared sequence similarity, not formal taxonomic designations. The boundaries of OTUs can be adjusted depending on the chosen similarity threshold and the level of resolution required for the study.
Question 5: What are the limitations of using OTUs?
OTUs can be limited by the chosen sequence similarity cut-off. A high similarity threshold may group distantly related organisms, while a low threshold might artificially increase the number of OTUs, diluting the significance of observed patterns. Furthermore, OTUs may not perfectly reflect the underlying species diversity and evolutionary relationships within a microbial community.
In summary, OTUs provide a valuable framework for analyzing microbial diversity and community structure. However, researchers should acknowledge their limitations and interpret data in light of the chosen operational criteria.
Moving forward, understanding the applications and limitations of OTUs will be crucial for conducting and interpreting microbial community studies accurately and effectively.
Conclusion
Operational taxonomic units (OTUs) have emerged as a powerful tool for characterizing and understanding microbial communities. This analysis highlights the multifaceted role of OTUs in ecological research, encompassing their application in assessing microbial diversity, analyzing community structure, and evaluating the impact of environmental factors. The use of sequence-based identification, particularly 16S rRNA gene sequencing, has been pivotal in defining OTUs, leading to a more comprehensive understanding of microbial evolutionary relationships. Comparative genomic analyses further illuminate the functional potential and adaptive strategies of microbial populations within identified OTUs. Consequently, the study of ecological relationships between OTUs reveals critical insights into the complex interactions within microbial communities, providing a framework for understanding ecosystem function. The practical implications of this knowledge extend to environmental monitoring, agricultural practices, and medical research. Despite the challenges inherent in species delineation and operational definitions, OTUs remain indispensable for unlocking the intricacies of microbial ecosystems.
The ongoing development and refinement of computational tools and analytical techniques are crucial for addressing the limitations of OTU-based analysis, particularly in resolving phylogenetic ambiguities. Future research should focus on integrating OTU data with other molecular and phenotypic data to achieve a more comprehensive understanding of microbial diversity and its influence on environmental processes. This integrated approach will further solidify the significance of OTUs in unraveling the complex tapestry of microbial life and its profound impact on Earth's ecosystems, human health, and the environment. Continued exploration will ultimately enhance our capability to effectively manage and conserve these vital components of our planet's biological systems.


Detail Author:
- Name : Mossie Wiegand
- Username : gschmidt
- Email : jacobson.isai@feil.biz
- Birthdate : 1973-10-24
- Address : 141 Tessie Orchard Nicoburgh, GA 78613-3853
- Phone : (484) 605-4105
- Company : Walsh, Wilkinson and Wolff
- Job : Bartender
- Bio : Ut quaerat blanditiis quia quia non officiis. Consequatur cupiditate sed perspiciatis enim beatae. Non dolores unde vero est aliquam.
Socials
twitter:
- url : https://twitter.com/sister_brown
- username : sister_brown
- bio : Labore quam omnis qui saepe dolorem natus quidem. Autem earum quos at ut rerum eum. Aut et fugit sed voluptas.
- followers : 5041
- following : 223
facebook:
- url : https://facebook.com/brown2013
- username : brown2013
- bio : Est nulla quaerat molestiae autem porro.
- followers : 5193
- following : 2318
instagram:
- url : https://instagram.com/sister_brown
- username : sister_brown
- bio : Enim quis placeat fuga. Officiis labore dolor in voluptatem sit dicta nemo.
- followers : 5981
- following : 2878
tiktok:
- url : https://tiktok.com/@brown1986
- username : brown1986
- bio : Et ea et eos explicabo. Sequi et et iusto reprehenderit eligendi facilis.
- followers : 3803
- following : 2861